Final Results
Visualizing Clonal Trees
Advisor: Eric Alexander
Times: Fall-Winter 1,2c
Background
In human biology, tumors result from an evolutionary process. Cells descending from a common ancestor accumulate genetic mutations that, in certain combinations, lead to uncontrolled cell growth and cancer. Knowing how a tumor evolved can be valuable for understanding why tumors develop, which may also lead to improved diagnosis and treatments.To this end, biologists describe the phylogenetic history of tumors using what are called clonal trees, which can be inferred from DNA sequencing data, though this area is still very much in development.
While some of the ongoing work on this topic is algorithmic (i.e., refining the algorithms by which these trees can be inferred), there is also a need for better tools for visualizing these trees. Existing techniques often rely on the standard node-link representations of trees that you’ve seen in Data Structures, as well as occasionally on less common techniques like “fish plots” and “tree maps” (see below). While each of these techniques has its strengths, they often suppress or fail to highlight data that is important to researchers.
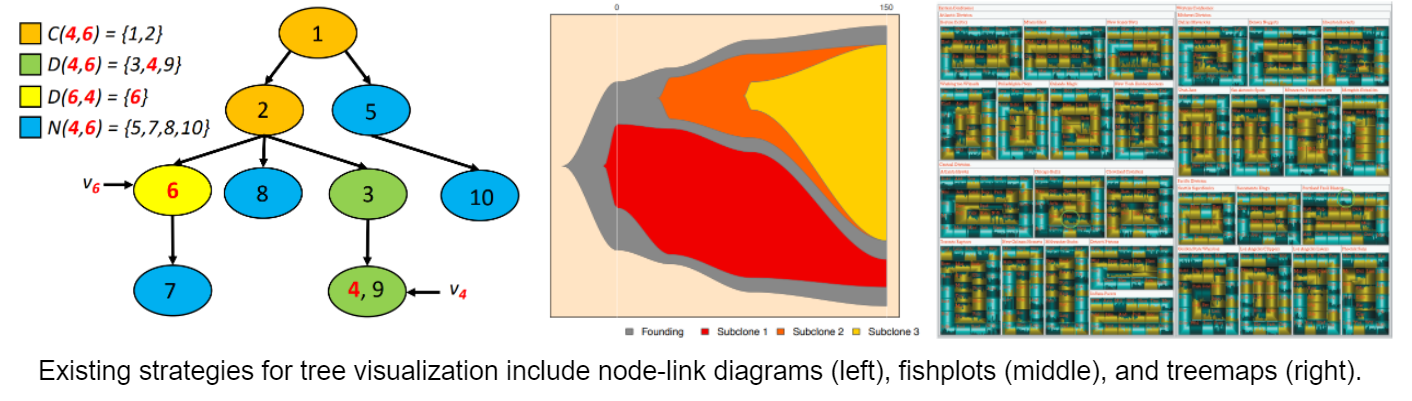
This project will have you design and implement a new tool for visualizing clonal trees that incorporates current best practices in data visualizations and seeks to meet the needs of researchers in this vibrant field.
The project
For this project, you will build an interactive tool to visualize clonal trees that have been inferred from DNA sequencing data. As part of the design of this tool, you will focus on two primary goals:
- Visualization of uncertainty and provenance. Inference is an inherently probabilistic and uncertain process. While there are metrics for describing that uncertainty, and how much a particular tree is supported by the underlying data, such information is rarely incorporated into visualization tools of this kind, which limits their usefulness in making clinical decisions.
- Affording visual comparison. While there are a plethora of techniques for visualizing a single tree (see here for a visual bibliography of many!), comparing trees–for instance, to better understand which of a variety of possible evolutions is most likely–is typically quite difficult.
You will start out with a review of existing literature in the realms of tree visualization and uncertainty visualization, along with relevant work on clonal trees specifically. You will then be tasked with carrying out a thorough design process, including contextual analysis of the problem domain, iterative prototyping and implementation, and usability testing. You will have the opportunity to work with Professor Layla Oesper and her research students to hone a tool that meets the needs of end users.
Deliverables
The deliverables for this project will be the code for the tool that you build, along with a write-up justifying its design.
Recommended experience
Experience in bioinformatics or data visualization (either through courses offered in the CS department, courses in other departments, or outside experiences) is a plus. CS 252 (Algorithms) and/or CS 257 (Software Design) may be valuable, as well. However, these different backgrounds need not be contained within any one person. Hopefully we will build a team with a variety of such experiences, and no single one or combination of these is required.