Final Results
Interactive Virtual Biology Labs
Advisor: Anya Vostinar
Background
Think back to the topics in class that you had the easiest time remembering. A lot of them were probably topics that you were able to interact with somehow, either by programming the concept directly or interacting with an example somehow. You might have even taken a science class that had a lab component, where you went in and performed your own experiments and observations. This way of learning, called active learning, it has been widely shown to be very effective at helping students deeply learn material in a way that they wouldn’t be able to by only reading a textbook or listening to a lecture. In particular, active learning has been shown to disproportionately help students from underprivileged and underrepresented backgrounds succeed, making it an issue of DEI (Haak et al., 2011, Theobald et al., 2016).
Despite the benefits of active learning, there are challenges to implementing it via traditional biology labs: 1) they are time consuming and expensive, 2) some concepts can’t be demonstrated due to temporal and spatial constraints, 3) there are situations where people can’t safely be in the same room together for long periods of time (like a pandemic). One strategy that has boomed in the last year due to necessity are browser-based virtual labs.
Two specific biological concepts that are important but difficult to demonstrate in traditional lab settings are evolution and endosymbiosis. Endosymbiosis is the biological concept of a symbiont (either a beneficial symbiont or a parasite) that lives inside of its host. The human gut microbiome is an example of a whole lot of endosymbionts that interact in complicated and interesting ways. My research software, Symbulation, models the evolution of endosymbiosis and has a basic browser-based GUI currently:
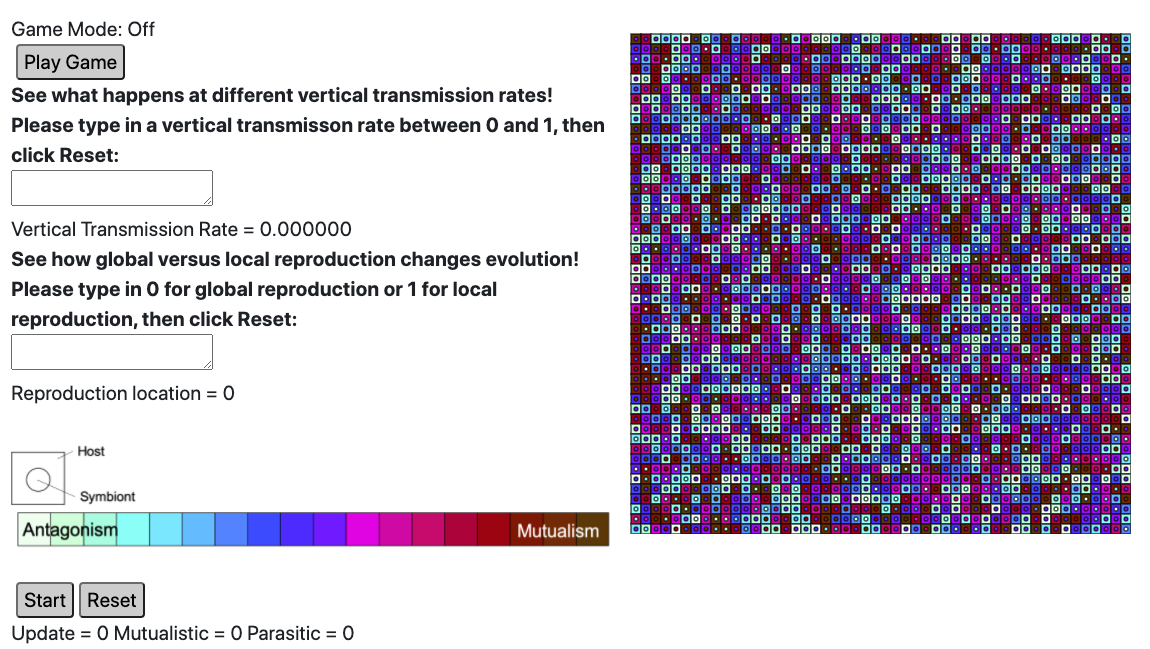
Your task in this comps project will be to add an educational lab to the existing GUI so that it walks users through relevant biological concepts and enables them to gather data relevant to their hypotheses.
The project
In this project, you’ll explore the design space of interactive virtual labs for biological concepts and build on the Symbulation software to create a virtual lab focused on the evolution of endosymbiosis. In particular, you will:
- Study the existing literature on virtual biology labs and do background reading on evolution and endosymbiosis.
- Submit an Institutional Review Board proposal to conduct a user study.
- Design a virtual lab built on top of Symbulation, using the open-source platform Empirical based on consultation with the Carleton Biology department.
- Conduct a user study to determine how effective your lab is at teaching the concepts.
Deliverables
The deliverables for this project will be the virtual lab code, ideally deployed to the Symbulation website and a report on the user study findings.
Recommended experience
This project uses the Symbulation and Empirical platforms that are written in C++ and JavaScript. Therefore, Software Design (CS 257) will be helpful on the front-end development and C or C++ experience will be helpful on the back-end development. In addition, biology coursework would be helpful but not required.
References/inspiration
Haak, D. C., HilleRisLambers, J., Pitre, E., & Freeman, S. (2011). Increased structure and active learning reduce the achievement gap in introductory biology. Science, 332 (6034), 1213–1216.
Theobald, E. J., Hill, M. J., Tran, E., Agrawal, S., Arroyo, E. N., Behling, S., Chambwe, N., Cintron, D. L., Cooper, J. D., Dunster, G., et al. (2020). Active learning narrows achievement gaps for underrepresented students in undergraduate science, technology, engineering, and math. Proceedings of the National Academy of Sciences, 117 (12), 6476–6483.
Avida-ED - similar software that has a virtual lab for evolution, but does not support endosymbiosis (yet, your work could end up influencing this project!)
Symbulation - my research software that you’ll build off of
Empirical - the platform Symbulation is built on, see the “Built With Empirical Gallery” for examples of what Empirical can support